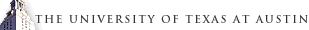PyAMFF: Python Atom-Centered Machine Learning Force Field
In recent years, machine learning algorithms have been widely used for the construction of force fields with accuracy of ab initio methods and efficiency of classical force fields. We developed a python-based atom-centered machine-learning force field (PyAMFF) package to provide a simple and efficient platform for fitting and using machine learning force fields by the implementation of an atomic-centered neural-network algorithm with Behler-Parrinello symmetry functions as structural fingerprints.
The following three features are included in PyAMFF: (1) integrated Fortran modules for fast fingerprint calculations and python modules for user-friendly integration through scripts and facile extension of future algorithms; (2) a pure Fortran backend to interface with the software including the long-timescale dynamic simulation package EON, enabling both molecular-dynamic simulations and adaptive kinetic Monte Carlo simulations with machine-learning force fields; and (3) integration with the Atomic Simulation Environment package for active learning and ML-based algorithm development.
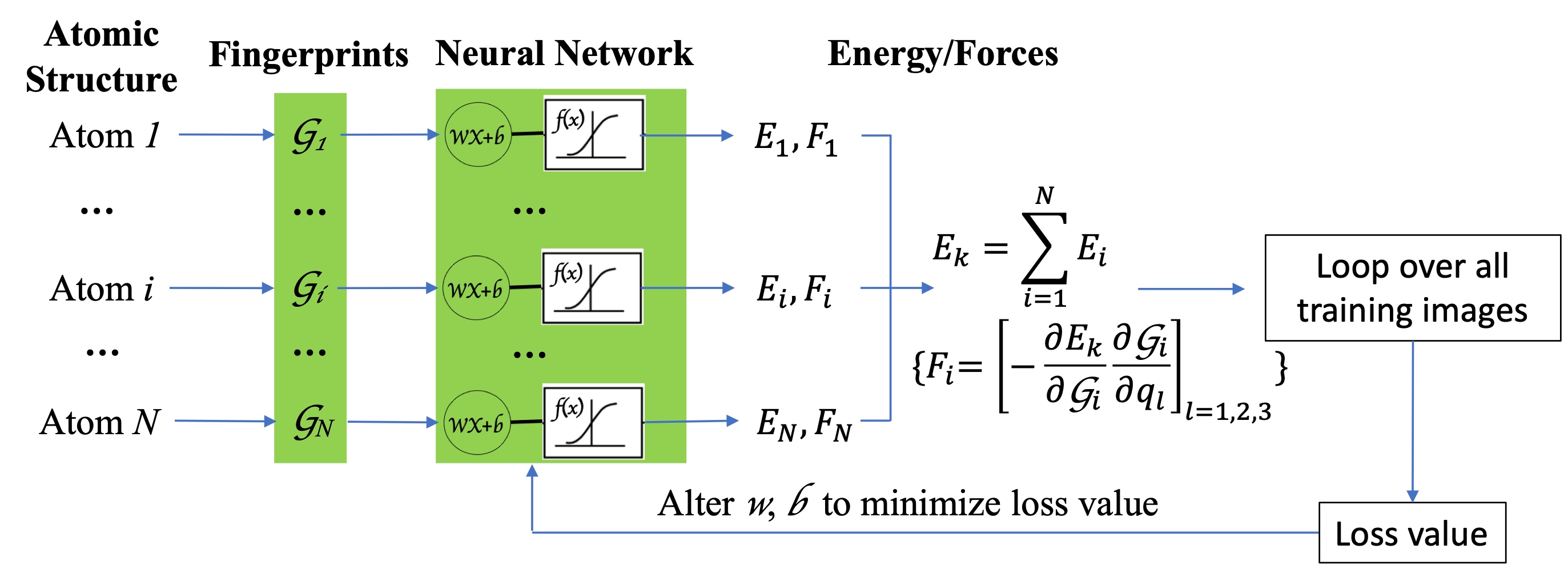
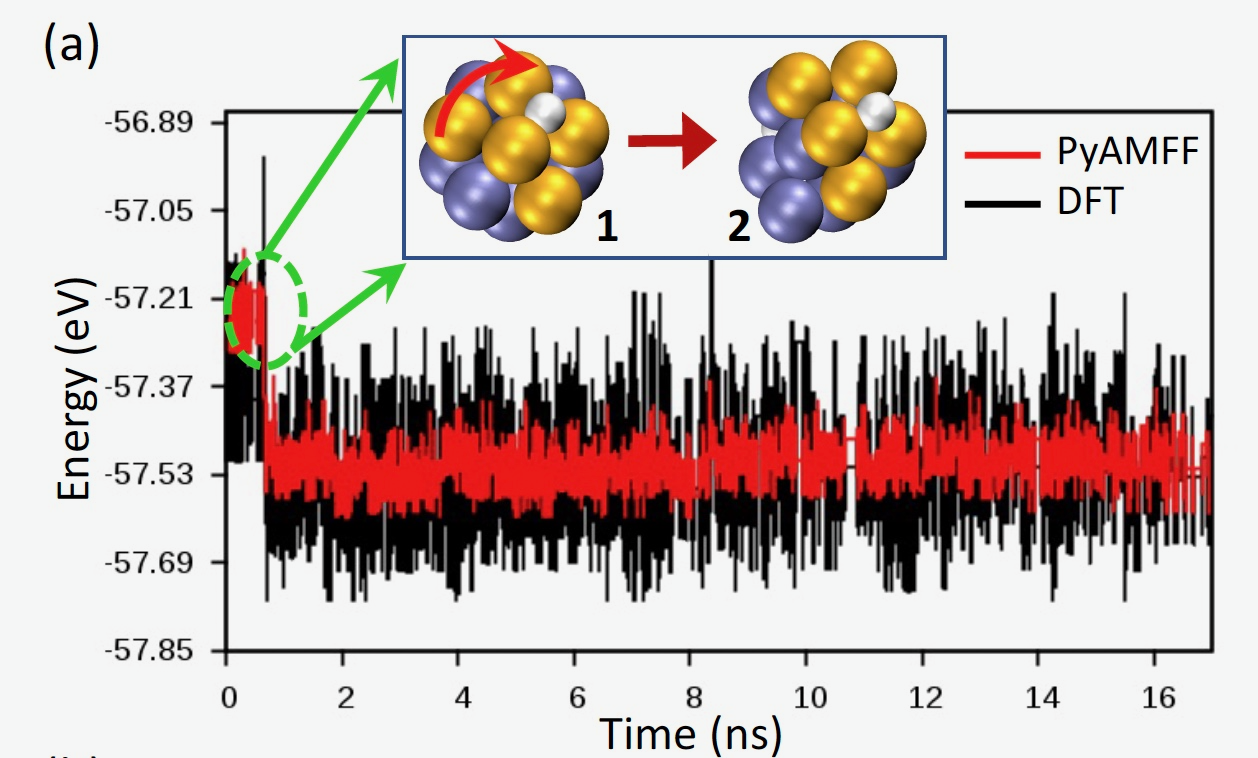
We performed AKMC simulations to compute the dynamical evolution of a Pd13H2 nanoparticle at 300 K, using the PyAMFF force field. Thanks to improved computation efficiency of fingerprints and their derivatives, the time scale of the dynamic simulation reaches 17 ns, much longer than that reported (100 ps) in Ref. The nanoparticle undergoes a structural transition accompanied by diffusion of the hydrogen atoms on the surface. As shown in Figure a, at ~0.6 ns, the rotation of a five-Pd motif (highlighted in gold) leads to a transition from an icosahedral structure to a deformed structure. Upon that, the system undergoes several metastable states (blue lines in Figure b) and transitions to a more stable state (red lines in Figure b). In such a state, the nanoparticle entails a chiral Pd structure as shown in inset images of Figure b. The chiral structures (3 and 4) correspond to the Type 4 and 5 isomers reported in Ref.
Fingerprint optimization for atom-centered machine learning force field
We have worked on developing simple schemes of selecting fingerprints as input for neural network force-field to best represent the atomic interactions. Selection of fingerprints based on radial and angular distribution functions have been shown to the symmetry functions which can characterize interactions correctly and develop accurate force-fields for studying potential energy surfaces. An example of LJ7 is shown here we a neural network based force-field was developed by selecting fingerprints from radial distribution function and the accuracy of the machine learning model was compared for different sets of fingerprints.
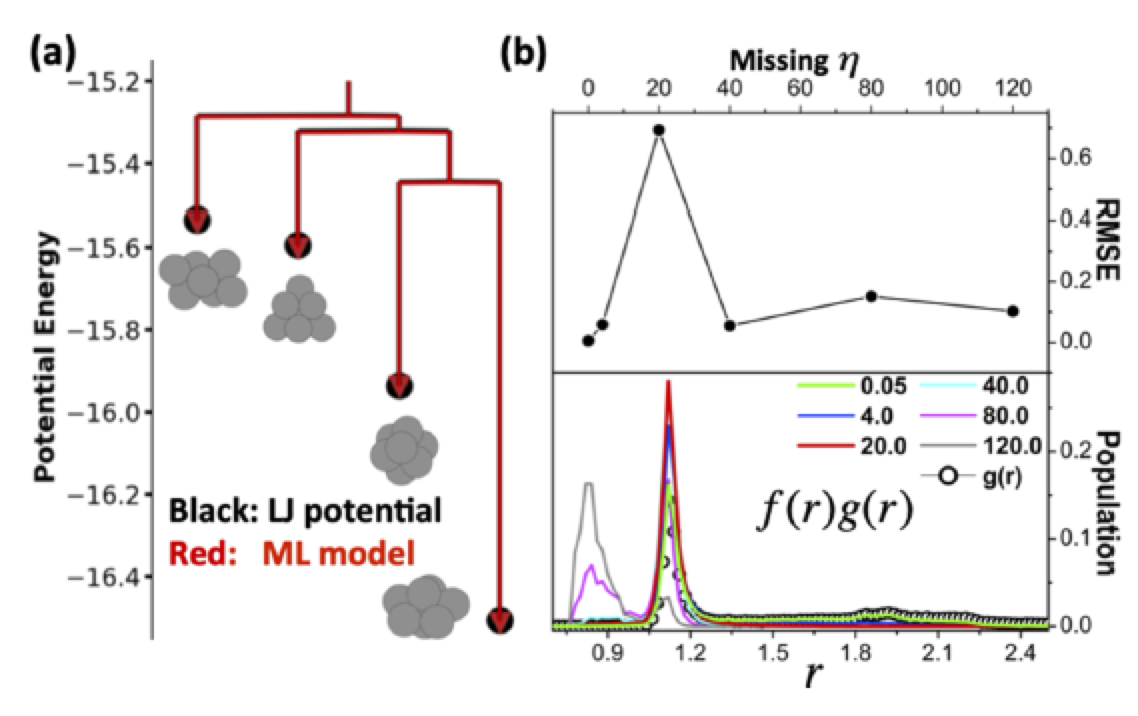
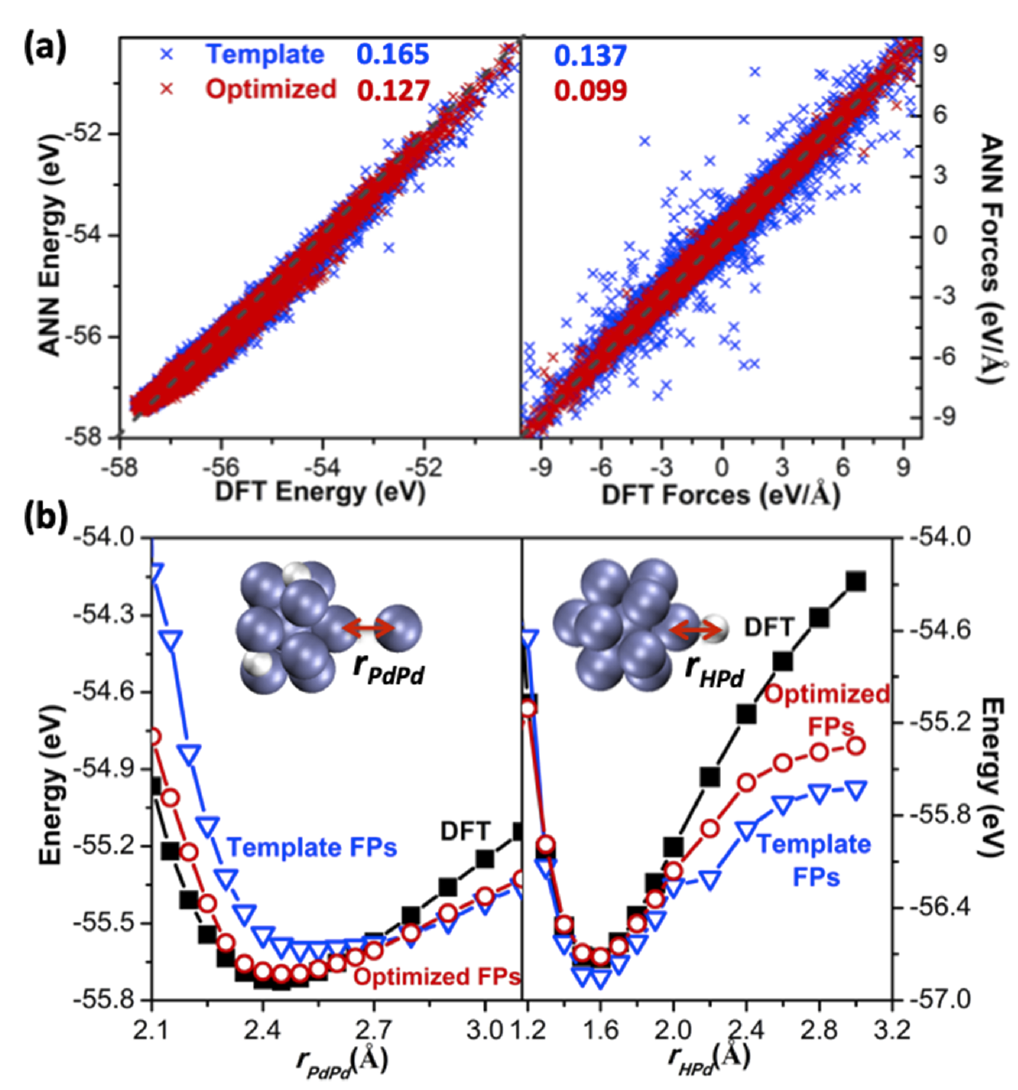
Developing an optimized set of fingerprints or symmetry functions can reduce the complexity of the input features needed to describe the force-field which is supposed to represent the potential energy surface of a given system. By carefully selecting fingerprints, can correctly capture the atomic interactions which resemble the properties of a true potential energy surface. The example shown below compares the performance of a trained neural network force-field with DFT where symmetry functions were optimized using the method shown above for the LJ7 case and without optimizing symmetry function (template fingerprints). We are constantly working on developing more analysis tools which can help in selecting representative symmetry functions, training structures and hyperparameters which can be useful for the community to develop generalized force-fields.
References
234
L. Li, H. Li, I. Seymour, L. Koziol, and G. Henkelman,
Pair-distribution-function Guided Optimization of Fingerprints for Atom-centered Neural Network Potentials,
J. Chem. Phys. 152, 224102 (2020).
DOI